Research projects and themes
Models for genetic variation
Using a cluster-based model for haplotype variation,  I develop and maintain the software package fastPHASE (Scheet & Stephens, 2006) for genotype imputation and
haplotype reconstruction. In Scheet & Stephens (2008),
I develop and maintain the software package fastPHASE (Scheet & Stephens, 2006) for genotype imputation and
haplotype reconstruction. In Scheet & Stephens (2008), 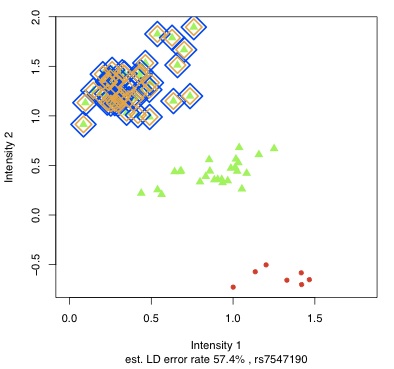 I extended this model to identify and correct genotyping errors. This model created
a framework for dealing with arbitrary data types and provides a natural way to improve resolution of genotypes in low-coverage next-generation
sequencing data. With my colleauge, Yongtao Guan, I am further developing models for genotypes and haplotypes in complex and structured populations.
(This work is supported from a grant to P. Scheet from the NIH.)
I extended this model to identify and correct genotyping errors. This model created
a framework for dealing with arbitrary data types and provides a natural way to improve resolution of genotypes in low-coverage next-generation
sequencing data. With my colleauge, Yongtao Guan, I am further developing models for genotypes and haplotypes in complex and structured populations.
(This work is supported from a grant to P. Scheet from the NIH.)
Genomic heterogeneity and diagnostics
 We have recently developed methods for profiling allelic imbalance in samples with ultra-low levels of tumor purity. These methods have natural applications for diagnostics and studying secondary and tertiary clones in heterogeneous cancer genomes.
We have recently developed methods for profiling allelic imbalance in samples with ultra-low levels of tumor purity. These methods have natural applications for diagnostics and studying secondary and tertiary clones in heterogeneous cancer genomes.
Genetic and genomic determinants of behavioral trajectories in twins
We are collaborating with Drs. Dorret Boomsma (Vrije Universiteit, Amsterdam), James Hudziak (Univ. of Vermont), and Gareth Davies (Univ. of South Dakota) to analyze over 9,000 GWA samples from 4,000 individuals in over 1,000 families. We are interested in how SNP and copy number variation affect behavior. The data come from the Netherlands Twin Registry and the study is funded by a Grand Opportunity ("GO") grant from the NIMH.
Pharmacogenomics and Survivorship
PAAR4Kids is a study of the pharmacogenetics/genomics of anticancer agents. In this study, we focus on childhood acute lymphoblastic leukemia ("ALL"). The project is led by Mary Relling of St. Jude Children's Research Hospital. Together with Gary Rosner, I provide statistical and statistical genetics expertise for aspects of the project. In addition we are using next-generation sequencing to detect germline risk alleles predisposing to adverse reactions to treatment in survivors of childhood ALL. This project is supported by a grant from the NIGMS and NCI.
Genotype imputation in diverse human populations
With Noah Rosenberg and Sebastian Zoellner at the University of Michigan, we are developing novel approaches to analyze genetic data from diverse and admixed populations, 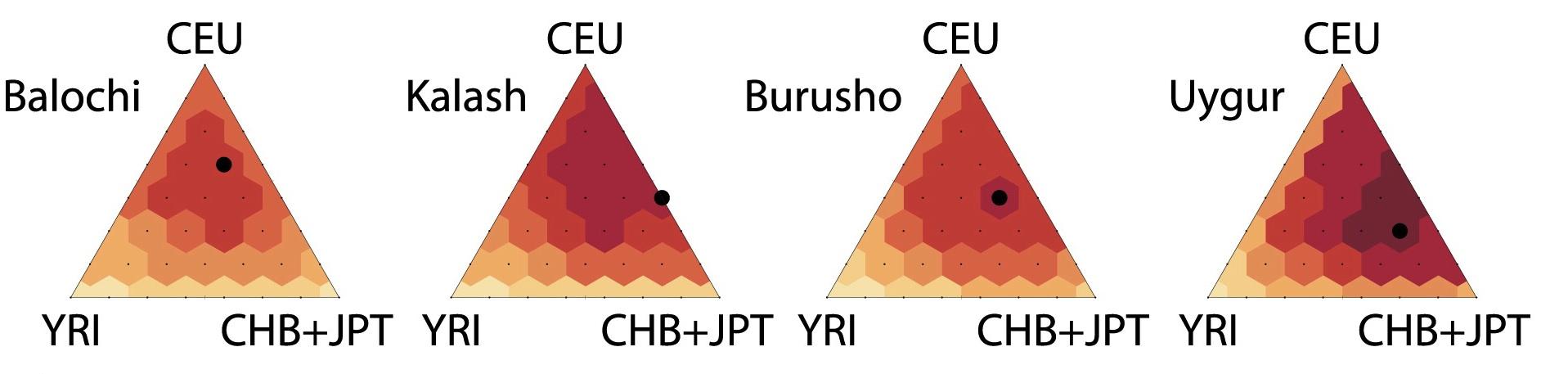 with a focus on imputing genotypes and rare genetic variation. In Huang
with a focus on imputing genotypes and rare genetic variation. In Huang
Genomic variation in CLL
With Lynne Abruzzo and Kevin Coombes and colleagues here at M. D. Anderson, we are helping to analyze genome-wide data of copy number and SNP variation in samples from patients with CLL. We are developing methods to detect loss of heterozygosity (LOH) in SNP data from unpaired tumor samples.
Genetic architecture of HLA
With our colleagues in rheumatology at UT Houston (John Reveille, Xiaodong Zhou, and Maureen Mays), we are fine-mapping the HLA region for association with Systemic Sclerosis and Ankylosing Spondylitis. (NIH; PI: Zhou)